DLinear model validation using offline batch inference#
This tutorial demonstrates how to perform batch inference using the DLinear model and Ray Data. The process involves loading the model checkpoint, preparing the test data, running inference in batches, and evaluating the performance.
Note that this notebook requires the pre-trained model artifacts that the previous “Distributed training of a DLinear time-series model” notebook generates.
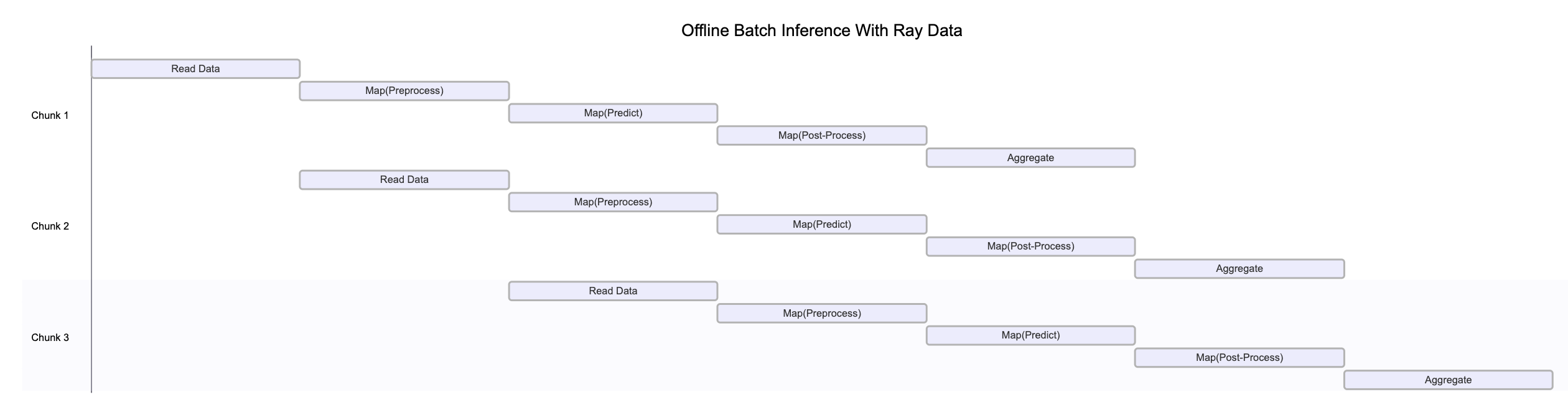
The preceding figure illustrates how different blocks of data process concurrently at various stages of the pipeline. This parallel execution maximizes resource utilization and throughput.
Note that this diagram is a simplification for various reasons:
Only one worker processes each data pipeline stage
Backpressure mechanisms may throttle upstream operators to prevent overwhelming downstream stages
Dynamic repartitioning often occurs as data moves through the pipeline, changing block counts and sizes
Available resources change as the cluster autoscales
System failures may disrupt the clean sequential flow shown in the diagram
❌ Traditional batch execution, non-streaming like Spark without pipelining or SageMaker Batch Transform:
Reads the entire dataset into memory or a persistent intermediate format
Only then starts applying transformations, such as
.map,.filter, etc.Higher memory pressure and startup latency
✅ Streaming execution with Ray Data:
Starts processing blocks as they load, without waiting for the entire dataset
Reduces memory footprint, preventing out-of-memory errors, and speeds up time to first output
Increases resource utilization by reducing idle time
Enables online-style inference pipelines with minimal latency
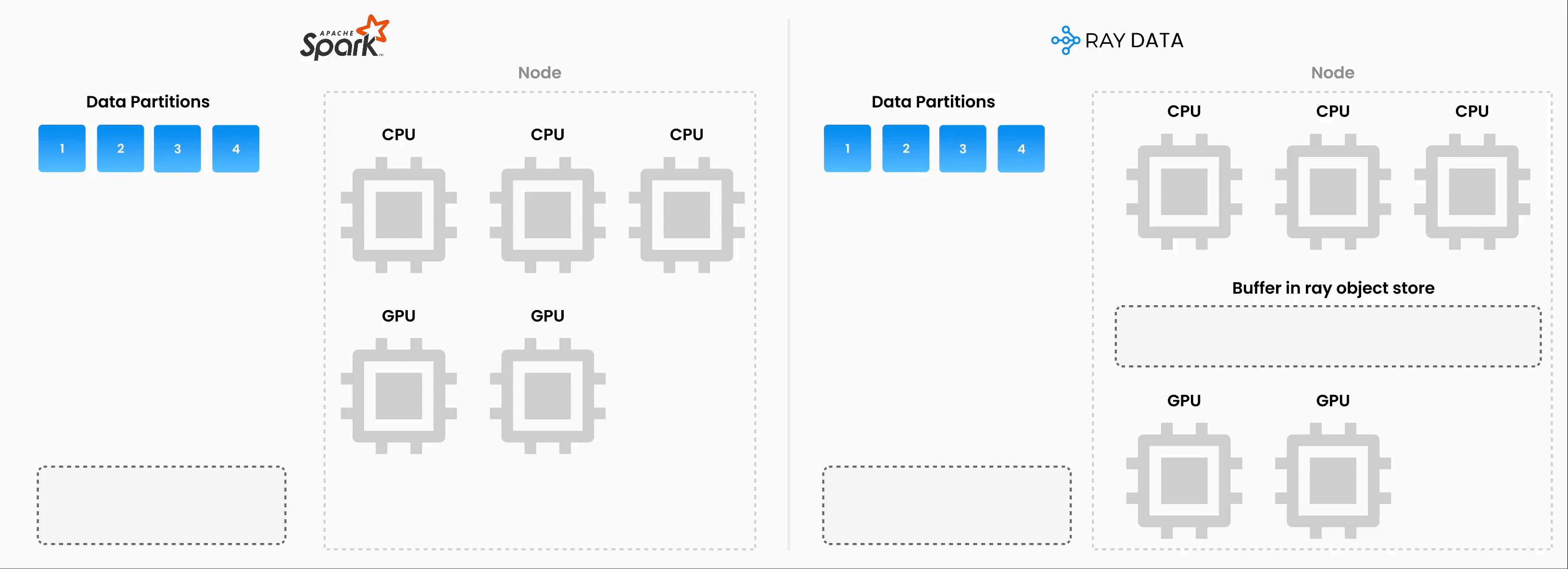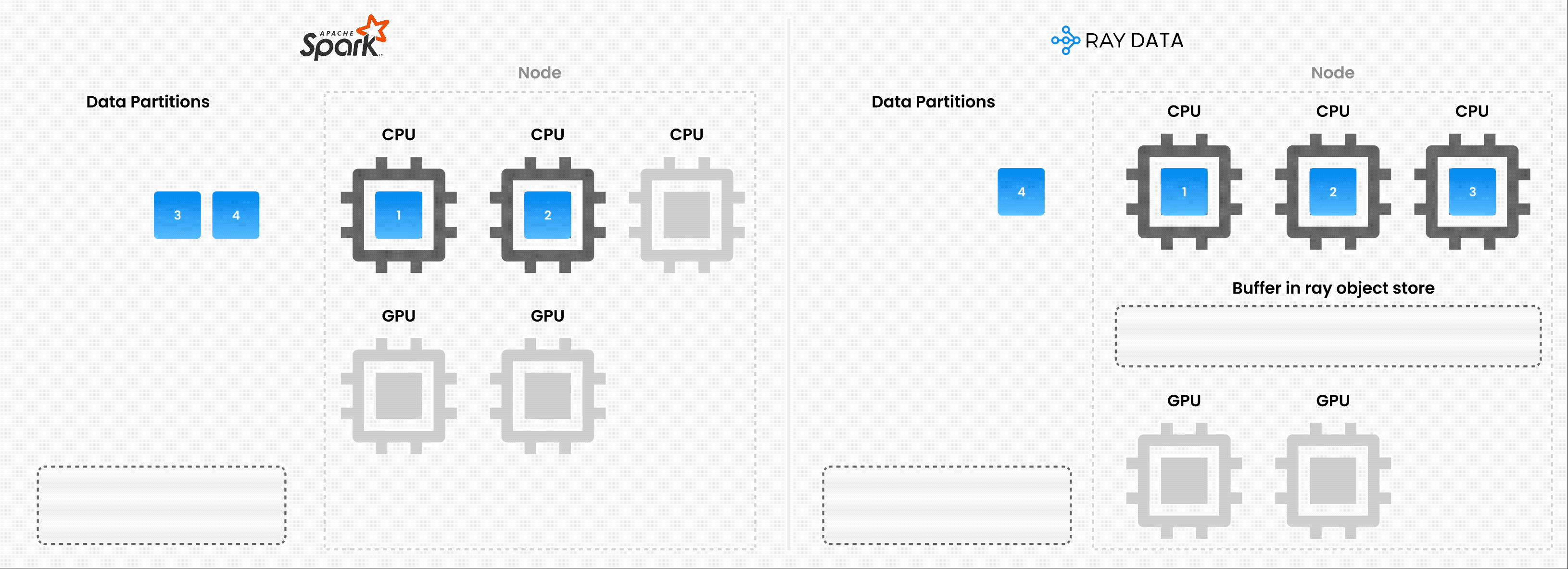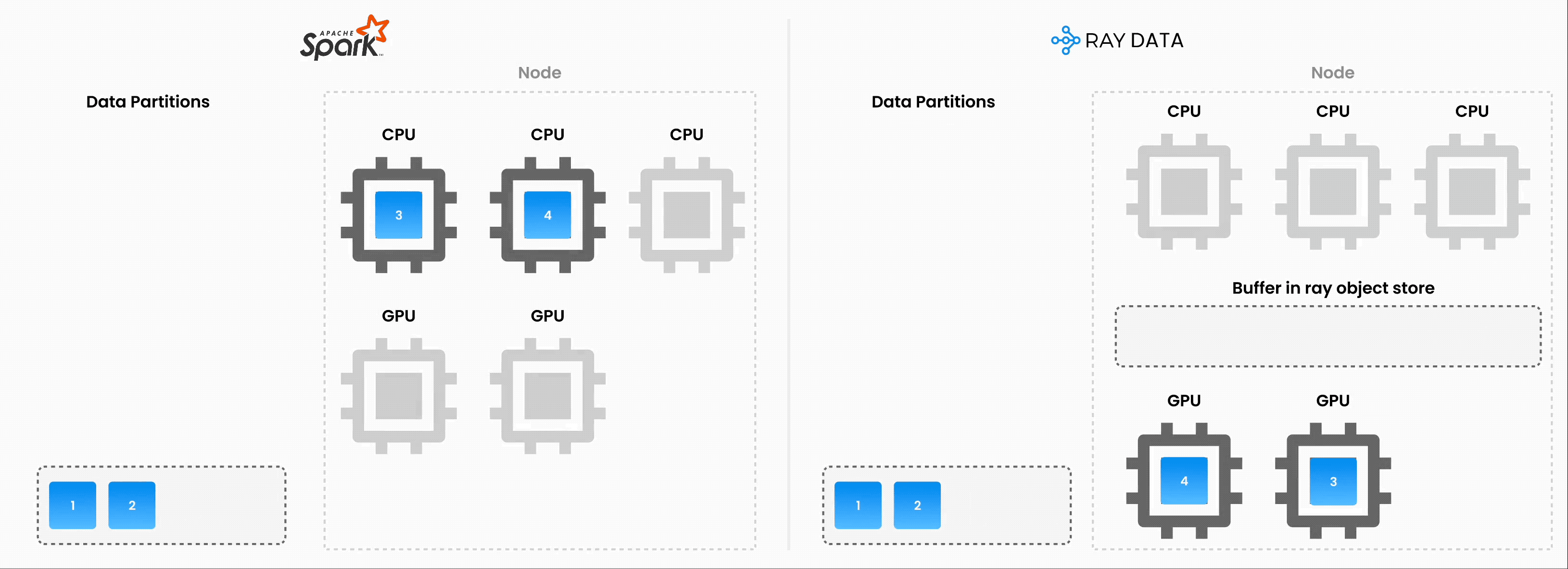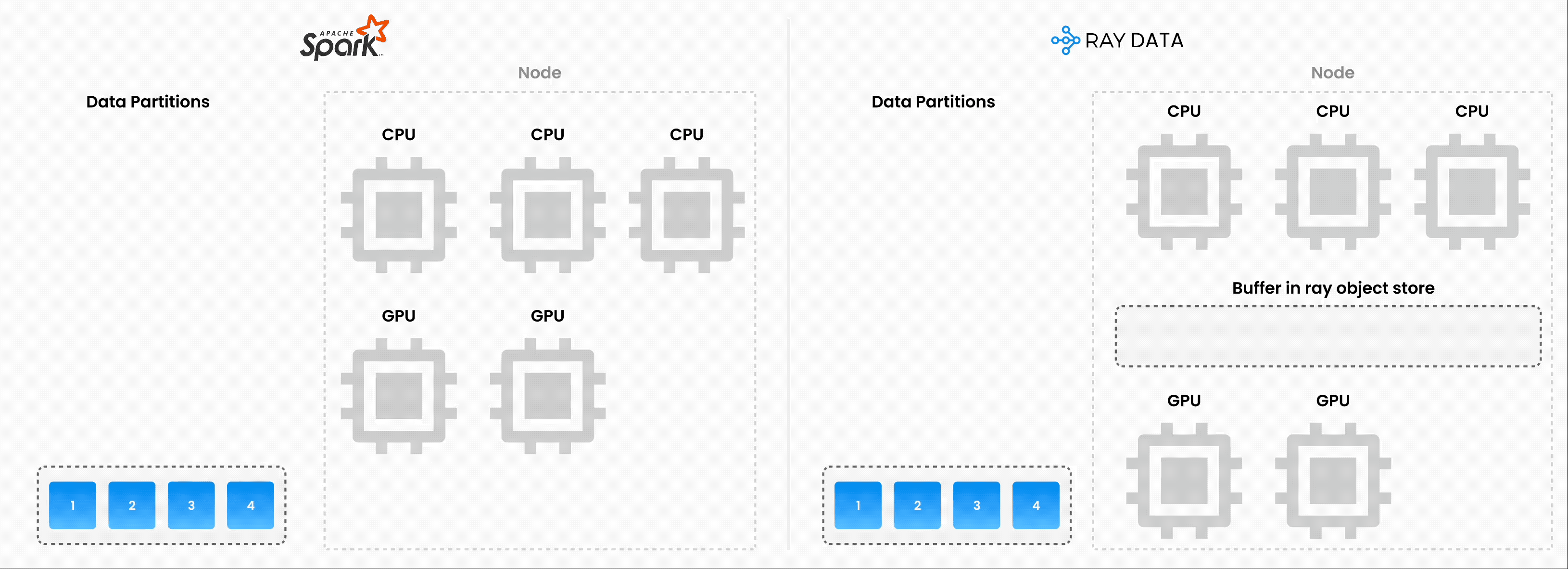
Note: Ray Data operates as batch processing with streaming execution rather than a real-time stream processing engine like Flink or Kafka Streams. This approach proves especially useful for iterative ML workloads, ETL pipelines, and preprocessing before training or inference. Ray typically delivers a 2-17x throughput improvement over solutions like Spark and SageMaker Batch Transform.
# Enable importing from e2e_timeseries module.
import os
import sys
sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd())))
Start by setting up the environment and imports:
import numpy as np
import ray
import torch
os.environ["RAY_TRAIN_V2_ENABLED"] = "1"
import e2e_timeseries
from e2e_timeseries.data_factory import data_provider
from e2e_timeseries.metrics import metric
from e2e_timeseries.model import DLinear
Initialize the Ray cluster with the e2e_timeseries module, so that newly spawned workers can import it.
ray.init(runtime_env={"py_modules": [e2e_timeseries]})
Next, set up the DLinear model configuration as well as job configuration:
# Load the best checkpoint path from the metadata file created in the training notebook.
best_checkpoint_metadata_fpath = os.path.join(
"/mnt/cluster_storage/checkpoints", "best_checkpoint_path.txt"
)
with open(best_checkpoint_metadata_fpath, "r") as f:
best_checkpoint_path = f.read().strip()
config = {
"checkpoint_path": best_checkpoint_path,
"num_data_workers": 1,
"features": "S",
"target": "OT",
"smoke_test": False,
"seq_len": 96,
"label_len": 48,
"pred_len": 96,
"individual": False,
"batch_size": 64,
"num_predictor_replicas": 4,
}
def _process_config(config: dict) -> dict:
"""Helper function to process and update configuration."""
# Configure encoder input size based on task type.
if config["features"] == "M" or config["features"] == "MS":
config["enc_in"] = 7 # ETTh1 has 7 features when multi-dimensional prediction is enabled
else:
config["enc_in"] = 1
# Ensure paths are absolute.
config["checkpoint_path"] = os.path.abspath(config["checkpoint_path"])
config["num_gpus_per_worker"] = 1.0
config["train_only"] = False # Load test subset
return config
# Set derived values.
config = _process_config(config)
Data ingest#
First, load the test dataset as a Ray Data Dataset. Use .show(1) to trigger the execution for a single row,
because Ray Data is lazily evaluates datasets.
ray.init(ignore_reinit_error=True)
print("Loading test data...")
ds = data_provider(config, flag="test")
ds.show(1)
This cell defines the Predictor class. It loads the trained DLinear model from a checkpoint and processes input batches to produce predictions. The call method performs inference on a given batch of NumPy arrays.
Ray Data’s actor-based processing enables loading the model weights and transferring them to GPU only once and reusing them across batches.
class Predictor:
"""Actor class for performing inference with the DLinear model."""
def __init__(self, checkpoint_path: str, config: dict):
self.config = config
self.device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Load model from checkpoint.
self.model = DLinear(config).float()
checkpoint = torch.load(checkpoint_path, map_location=self.device)
self.model.load_state_dict(checkpoint["model_state_dict"])
self.model.to(self.device)
self.model.eval()
def __call__(self, batch: dict[str, np.ndarray]) -> dict:
"""Process a batch of data for inference (numpy batch format)."""
# Convert input batch to tensor.
batch_x = torch.from_numpy(batch["x"]).float().to(self.device)
with torch.no_grad():
outputs = self.model(batch_x) # Shape (N, pred_len, features_out)
# Determine feature dimension based on config.
f_dim = -1 if self.config["features"] == "MS" else 0
outputs = outputs[:, -self.config["pred_len"] :, f_dim:]
outputs_np = outputs.cpu().numpy()
# Extract the target part from the batch.
batch_y = batch["y"]
batch_y_target = batch_y[:, -self.config["pred_len"] :]
return {"predictions": outputs_np, "targets": batch_y_target}
ds = ds.map_batches(
Predictor,
fn_constructor_kwargs={"checkpoint_path": config["checkpoint_path"], "config": config},
batch_size=config["batch_size"],
concurrency=config["num_predictor_replicas"],
num_gpus=config["num_gpus_per_worker"],
batch_format="numpy",
)
Next, perform minor post-processing to get the results in the desired dimensions.
def postprocess_items(item: dict) -> dict:
# Squeeze singleton dimensions for predictions and targets if necessary.
if item["predictions"].shape[-1] == 1:
item["predictions"] = item["predictions"].squeeze(-1)
if item["targets"].shape[-1] == 1:
item["targets"] = item["targets"].squeeze(-1)
return item
ds = ds.map(postprocess_items)
Finally, execute all of these lazy steps and materialize them into memory using take_all():
# Trigger the lazy execution of the entire Ray pipeline.
all_results = ds.take_all()
Now that the results are in memory, calculate some validation metrics for the trained DLinear model.
# Concatenate predictions and targets from all batches.
all_predictions = np.concatenate([item["predictions"] for item in all_results], axis=0)
all_targets = np.concatenate([item["targets"] for item in all_results], axis=0)
# Compute evaluation metrics.
mae, mse, rmse, mape, mspe, rse = metric(all_predictions, all_targets)
print("\n--- Test Results ---")
print(f"MSE: {mse:.3f}")
print(f"MAE: {mae:.3f}")
print(f"RMSE: {rmse:.3f}")
print(f"MAPE: {mape:.3f}")
print(f"MSPE: {mspe:.3f}")
print(f"RSE: {rse:.3f}")
print("\nOffline inference finished!")

