Scalable online XGBoost inference with Ray Serve#
This tutorial launches an online service that:
deploys trained XGBoost model artifacts to generate predictions
autoscales based on real-time incoming traffic
covers observability and debugging around the service
Note that this notebook requires that you run the Distributed training of an XGBoost model tutorial to generate the pre-trained model artifacts that this tutorial fetches.
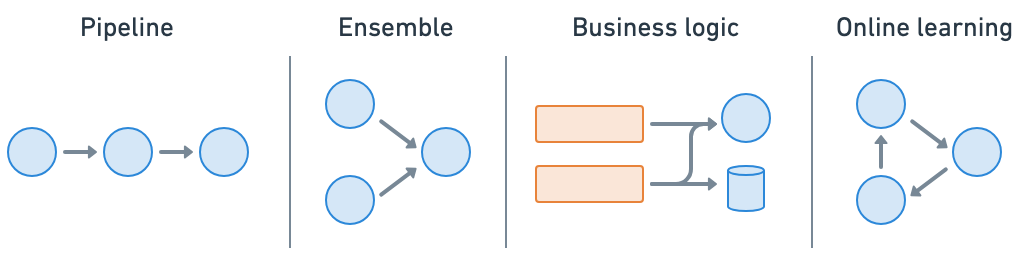
Ray Serve is a highly scalable and flexible model serving library for building online inference APIs. You can:
Wrap models and business logic as separate serve deployments and connect them together (pipeline, ensemble, etc.)
Avoid one large service that’s network and compute bounded and an inefficient use of resources
Utilize fractional heterogeneous resources, which isn’t possible with SageMaker, Vertex, KServe, etc., and horizontally scale, with
num_replicasAutoscale up and down based on traffic
Integrate with FastAPI and HTTP
Set up a gRPC service to build distributed systems and microservices
Enable dynamic batching based on batch size, time, etc.
Access a suite of utilities for serving LLMs that are inference-engine agnostic and have batteries-included support for LLM-specific features such as multi-LoRA support

%load_ext autoreload
%autoreload all
# Enable loading of the dist_xgboost module.
import os
import sys
sys.path.append(os.path.abspath(".."))
# Enable Ray Train v2.
os.environ["RAY_TRAIN_V2_ENABLED"] = "1"
# Now it's safe to import from ray.train.
import ray
import dist_xgboost
# Initialize Ray with the dist_xgboost package.
ray.init(runtime_env={"py_modules": [dist_xgboost]})
Loading the model#
Next, load the pre-trained preprocessor and XGBoost model from the MLflow registry as demonstrated in the validation notebook.
Creating a Ray Serve deployment#
Next, define the Ray Serve endpoint. Use a reusable class to avoid reloading the model and preprocessor for each request. The deployment supports both Pythonic and HTTP requests.
import pandas as pd
import xgboost
from ray import serve
from starlette.requests import Request
from dist_xgboost.data import load_model_and_preprocessor
@serve.deployment(num_replicas=2, max_ongoing_requests=25, ray_actor_options={"num_cpus": 2})
class XGBoostModel:
def __init__(self):
self.preprocessor, self.model = load_model_and_preprocessor()
@serve.batch(max_batch_size=16, batch_wait_timeout_s=0.1)
async def predict_batch(self, input_data: list[dict]) -> list[float]:
print(f"Batch size: {len(input_data)}")
# Convert list of dictionaries to DataFrame.
input_df = pd.DataFrame(input_data)
# Preprocess the input.
preprocessed_batch = self.preprocessor.transform_batch(input_df)
# Create DMatrix for prediction.
dmatrix = xgboost.DMatrix(preprocessed_batch)
# Get predictions.
predictions = self.model.predict(dmatrix)
return predictions.tolist()
async def __call__(self, request: Request):
# Parse the request body as JSON.
input_data = await request.json()
return await self.predict_batch(input_data)
Ray Serve makes it extremely easy to do model composition where you can compose multiple deployments containing ML models or business logic into a single application. You can independently scale even fractional resources, and configure each of the deployments.
Ensure that you don’t have any existing deployments first using serve.shutdown():
if "default" in serve.status().applications and serve.status().applications["default"].status == "RUNNING":
print("Shutting down existing serve application")
serve.shutdown()
2025-04-16 21:35:03,819 INFO worker.py:1660 -- Connecting to existing Ray cluster at address: 10.0.23.200:6379...
2025-04-16 21:35:03,828 INFO worker.py:1843 -- Connected to Ray cluster. View the dashboard at https://session-1kebpylz8tcjd34p4sv2h1f9tg.i.anyscaleuserdata.com
2025-04-16 21:35:03,833 INFO packaging.py:367 -- Pushing file package 'gcs://_ray_pkg_dbf2a602028d604b4b1f9474b353f0574c4a48ce.zip' (0.08MiB) to Ray cluster...
2025-04-16 21:35:03,834 INFO packaging.py:380 -- Successfully pushed file package 'gcs://_ray_pkg_dbf2a602028d604b4b1f9474b353f0574c4a48ce.zip'.
Now that you’ve defined the deployment, you can create a ray.serve.Application using the .bind() method:
# Define the app.
xgboost_model = XGBoostModel.bind()
Preparing test data#
Prepare some example data to test the deployment. Use a sample from the hold-out set:
sample_input = {
"mean radius": 14.9,
"mean texture": 22.53,
"mean perimeter": 102.1,
"mean area": 685.0,
"mean smoothness": 0.09947,
"mean compactness": 0.2225,
"mean concavity": 0.2733,
"mean concave points": 0.09711,
"mean symmetry": 0.2041,
"mean fractal dimension": 0.06898,
"radius error": 0.253,
"texture error": 0.8749,
"perimeter error": 3.466,
"area error": 24.19,
"smoothness error": 0.006965,
"compactness error": 0.06213,
"concavity error": 0.07926,
"concave points error": 0.02234,
"symmetry error": 0.01499,
"fractal dimension error": 0.005784,
"worst radius": 16.35,
"worst texture": 27.57,
"worst perimeter": 125.4,
"worst area": 832.7,
"worst smoothness": 0.1419,
"worst compactness": 0.709,
"worst concavity": 0.9019,
"worst concave points": 0.2475,
"worst symmetry": 0.2866,
"worst fractal dimension": 0.1155,
}
sample_target = 0 # Ground truth label
Running the service#
There are two ways to run a Ray Serve service:
Serve API: use the
serve runCLI command, likeserve run tutorial:xgboost_model.Pythonic API: use
ray.serve’sserve.runcommand, likeserve.run(xgboost_model).
This example uses the Pythonic API:
from ray.serve.handle import DeploymentHandle
handle: DeploymentHandle = serve.run(xgboost_model, name="xgboost-breast-cancer-classifier")
INFO 2025-04-16 21:35:08,246 serve 30790 -- Started Serve in namespace "serve".
INFO 2025-04-16 21:35:13,363 serve 30790 -- Application 'xgboost-breast-cancer-classifier' is ready at http://127.0.0.1:8000/.
(ProxyActor pid=31032) INFO 2025-04-16 21:35:08,167 proxy 10.0.23.200 -- Proxy starting on node dc30e171b93f61245644ba4d0147f8b27f64e9e1eaf34d1bb63c9c99 (HTTP port: 8000).
(ProxyActor pid=31032) INFO 2025-04-16 21:35:08,226 proxy 10.0.23.200 -- Got updated endpoints: {}.
(ServeController pid=30973) INFO 2025-04-16 21:35:08,307 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ProxyActor pid=31032) INFO 2025-04-16 21:35:08,310 proxy 10.0.23.200 -- Got updated endpoints: {Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier'): EndpointInfo(route='/', app_is_cross_language=False)}.
(ProxyActor pid=31032) INFO 2025-04-16 21:35:08,323 proxy 10.0.23.200 -- Started <ray.serve._private.router.SharedRouterLongPollClient object at 0x77864005ee70>.
(ServeController pid=30973) INFO 2025-04-16 21:35:08,411 controller 30973 -- Adding 2 replicas to Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier').
(ServeController pid=30973) INFO 2025-04-16 21:35:09,387 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ServeController pid=30973) INFO 2025-04-16 21:35:10,337 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ServeController pid=30973) INFO 2025-04-16 21:35:10,550 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ServeController pid=30973) INFO 2025-04-16 21:35:11,395 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ServeController pid=30973) INFO 2025-04-16 21:35:12,449 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ServeController pid=30973) INFO 2025-04-16 21:35:13,402 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ServeController pid=30973) INFO 2025-04-16 21:35:13,613 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
You should see some logs indicating that the service is running locally:
INFO 2025-04-09 14:06:55,760 serve 31684 -- Started Serve in namespace "serve".
INFO 2025-04-09 14:06:57,875 serve 31684 -- Application 'default' is ready at http://127.0.0.1:8000/.
You can also check whether it’s running using serve.status():
serve.status().applications["xgboost-breast-cancer-classifier"].status == "RUNNING"
True
Querying the service#
Using HTTP#
The most common way to query services is with an HTTP request. This request invokes the __call__ method defined earlier:
import requests
url = "http://127.0.0.1:8000/"
prediction = requests.post(url, json=sample_input).json()
print(f"Prediction: {prediction:.4f}")
print(f"Ground truth: {sample_target}")
Prediction: 0.0503
Ground truth: 0
This approach works for processing an individual query, but isn’t appropriate if you have many queries. Because requests.post is a blocking call, if you run it in a for loop you never benefit from Ray Serve’s dynamic batching.
Instead, you want to fire many requests concurrently using asynchronous requests and let Ray Serve buffer and batch process them. You can use this approach with aiohttp:
import asyncio
import aiohttp
async def fetch(session, url, data):
async with session.post(url, json=data) as response:
return await response.json()
async def fetch_all(requests: list):
async with aiohttp.ClientSession() as session:
tasks = [fetch(session, url, input_item) for input_item in requests]
responses = await asyncio.gather(*tasks)
return responses
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) Batch size: 1
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:13,834 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 0ddcd27d-d671-4365-b7e3-6e4cae856d9b -- POST / 200 117.8ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,352 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 aeb83339-359a-41e2-99c4-4ab06252d0b9 -- POST / 200 94.7ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,353 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 8c80adfd-2033-41d3-a718-aecbd5bcb996 -- POST / 200 93.9ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,354 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 7ed45f79-c665-4a17-94f7-6d02c56ab504 -- POST / 200 93.8ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,355 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 56fd016b-497a-43cc-b500-edafe878cda8 -- POST / 200 88.6ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,356 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 4910e208-d042-4fcb-aba9-330400fba538 -- POST / 200 85.5ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,356 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 b4999d9c-72fd-4bd2-aa9c-3c854ebe7457 -- POST / 200 84.7ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,358 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 04bc7c27-ae22-427f-8bee-c9dbc48a0b82 -- POST / 200 85.3ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,358 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 dcbbe5fa-d278-4568-a0fb-ea9347889990 -- POST / 200 84.3ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,359 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 22683613-16a5-479a-92bc-14f07dc317aa -- POST / 200 83.3ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,360 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 b773626c-8607-4572-bb87-8d8f80964de5 -- POST / 200 82.8ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,361 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 bceee2b4-ff30-4866-a300-7591e0cdc598 -- POST / 200 79.2ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,362 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 edaeb2f7-8de3-494d-8db0-8ebf2009acf7 -- POST / 200 74.7ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,362 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 09a38fe8-47d3-4c0e-8f5e-c312cded2c35 -- POST / 200 74.6ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,363 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 7f0d2f52-e59b-4f26-8931-61a1e9e4f988 -- POST / 200 72.9ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,363 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 269b045d-0b42-407d-a52f-7222cafce0d6 -- POST / 200 71.5ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) INFO 2025-04-16 21:35:14,364 xgboost-breast-cancer-classifier_XGBoostModel cxd4bxd1 98b7ef19-f5a1-4ab2-a71c-a2b7f6a6c1ad -- POST / 200 71.1ms
(ServeController pid=30973) INFO 2025-04-16 21:35:14,457 controller 30973 -- Deploying new version of Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier') (initial target replicas: 2).
(ProxyActor pid=5012, ip=10.0.240.129) INFO 2025-04-16 21:35:14,484 proxy 10.0.240.129 -- Proxy starting on node 9d22416ba66c129a3b66c96533eaa5455f7e882c37408b4fe7dc81f8 (HTTP port: 8000).
sample_input_list = [sample_input] * 100
# Notebook is already running an asyncio event loop in background, so use `await`.
# In other cases, you would use `asyncio.run(fetch_all(sample_input_list))`.
responses = await fetch_all(sample_input_list)
print(f"Finished processing {len(responses)} queries. Example result: {responses[0]}")
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) Batch size: 16
Finished processing 100 queries. Example result: 0.05025313049554825
(ProxyActor pid=5012, ip=10.0.240.129) INFO 2025-04-16 21:35:14,555 proxy 10.0.240.129 -- Got updated endpoints: {Deployment(name='XGBoostModel', app='xgboost-breast-cancer-classifier'): EndpointInfo(route='/', app_is_cross_language=False)}.
(ProxyActor pid=5012, ip=10.0.240.129) INFO 2025-04-16 21:35:14,576 proxy 10.0.240.129 -- Started <ray.serve._private.router.SharedRouterLongPollClient object at 0x7835f2b9acc0>.
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,619 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 24933cc1-07b4-4680-bb84-adcd54ff2de3 -- POST / 200 139.5ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,620 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 15167894-ceac-4464-bbb6-0556c8299d8a -- POST / 200 138.3ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,621 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x e4bb73d9-6b5b-4cd0-8dc0-5bbe5329c29e -- POST / 200 138.6ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,621 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 004be5f3-9ce7-4708-8579-31da77926491 -- POST / 200 94.1ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,621 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 233fc1bb-6486-4704-bf03-8599176e539c -- POST / 200 92.7ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,621 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x cd417685-cad4-4c9d-ab51-fcd33babe57c -- POST / 200 88.5ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,622 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 0ea1c55a-6722-4cb6-a9ab-9e0ffa156ef4 -- POST / 200 84.6ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,622 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 3315400d-9213-46ac-9abd-baa576c73107 -- POST / 200 77.9ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,622 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 25054e1f-e3e7-4106-910b-f6ba94f111be -- POST / 200 76.9ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,623 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x a0dbd826-c595-455f-8869-7c567c0dfac2 -- POST / 200 75.6ms
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) INFO 2025-04-16 21:35:14,623 xgboost-breast-cancer-classifier_XGBoostModel ep2o1d1x 136060ac-9705-49a5-b743-dc29164a3eee -- POST / 200 75.4ms
Using Python#
For a more direct Pythonic way to query the model, you can use the deployment handle:
response = await handle.predict_batch.remote(sample_input)
print(response)
INFO 2025-04-16 21:35:14,803 serve 30790 -- Started <ray.serve._private.router.SharedRouterLongPollClient object at 0x7156ffcf6d80>.
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4874, ip=10.0.240.129) Batch size: 11
(ServeReplica:xgboost-breast-cancer-classifier:XGBoostModel pid=4875, ip=10.0.240.129) Batch size: 1
0.05025313049554825
This approach is useful if you need to interact with the service from a different process in the same Ray Cluster. If you need to regenerate the serve handle, you can use serve.get_deployment_handle:
handle = serve.get_deployment_handle("XGBoostModel", "xgboost-breast-cancer-classifier")
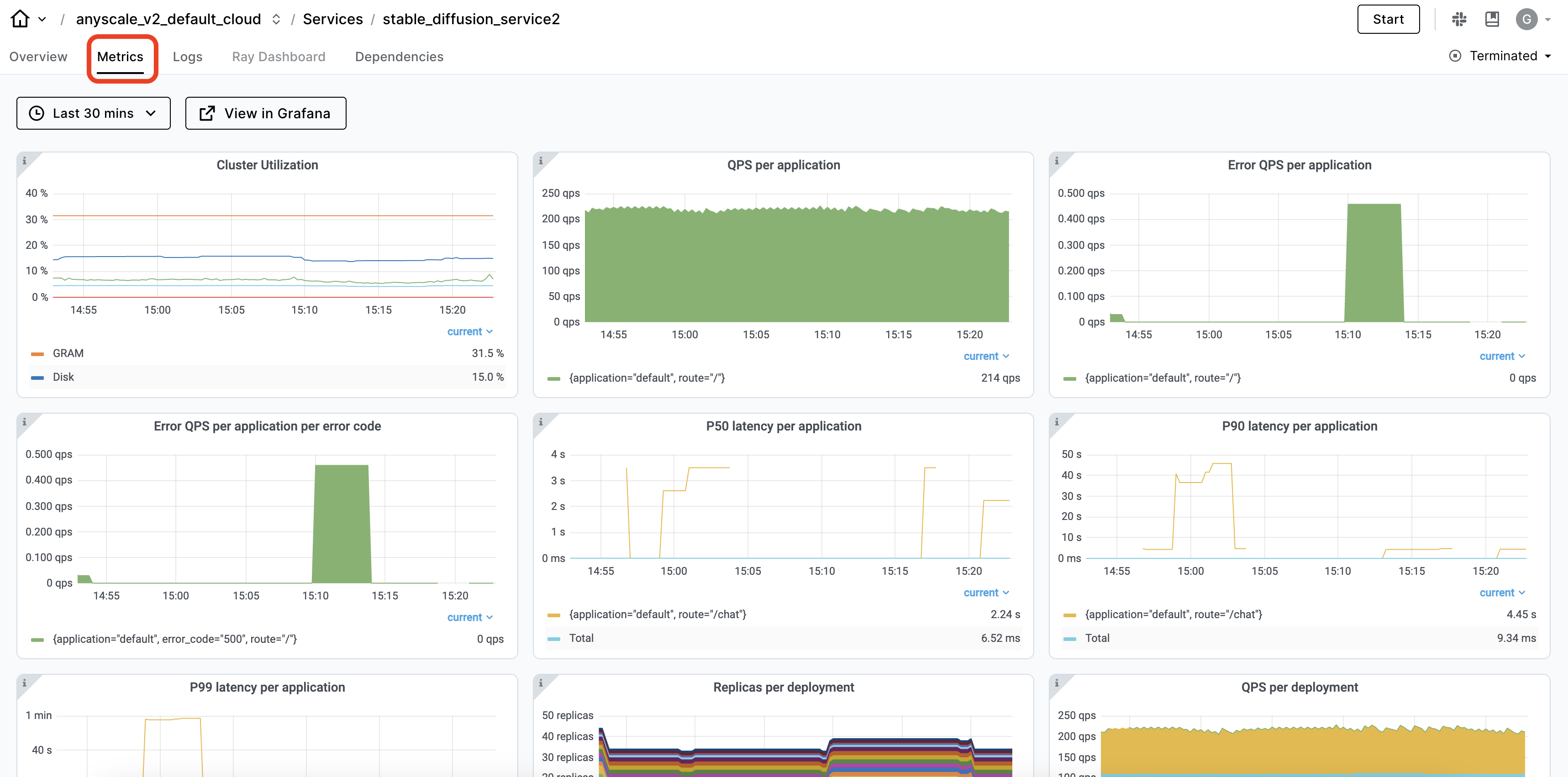
The Ray dashboard automatically captures observability for Ray Serve applications in the Serve view. You can view the service deployments and their replicas and time-series metrics about the service’s health.
# Shutdown service.
serve.shutdown()
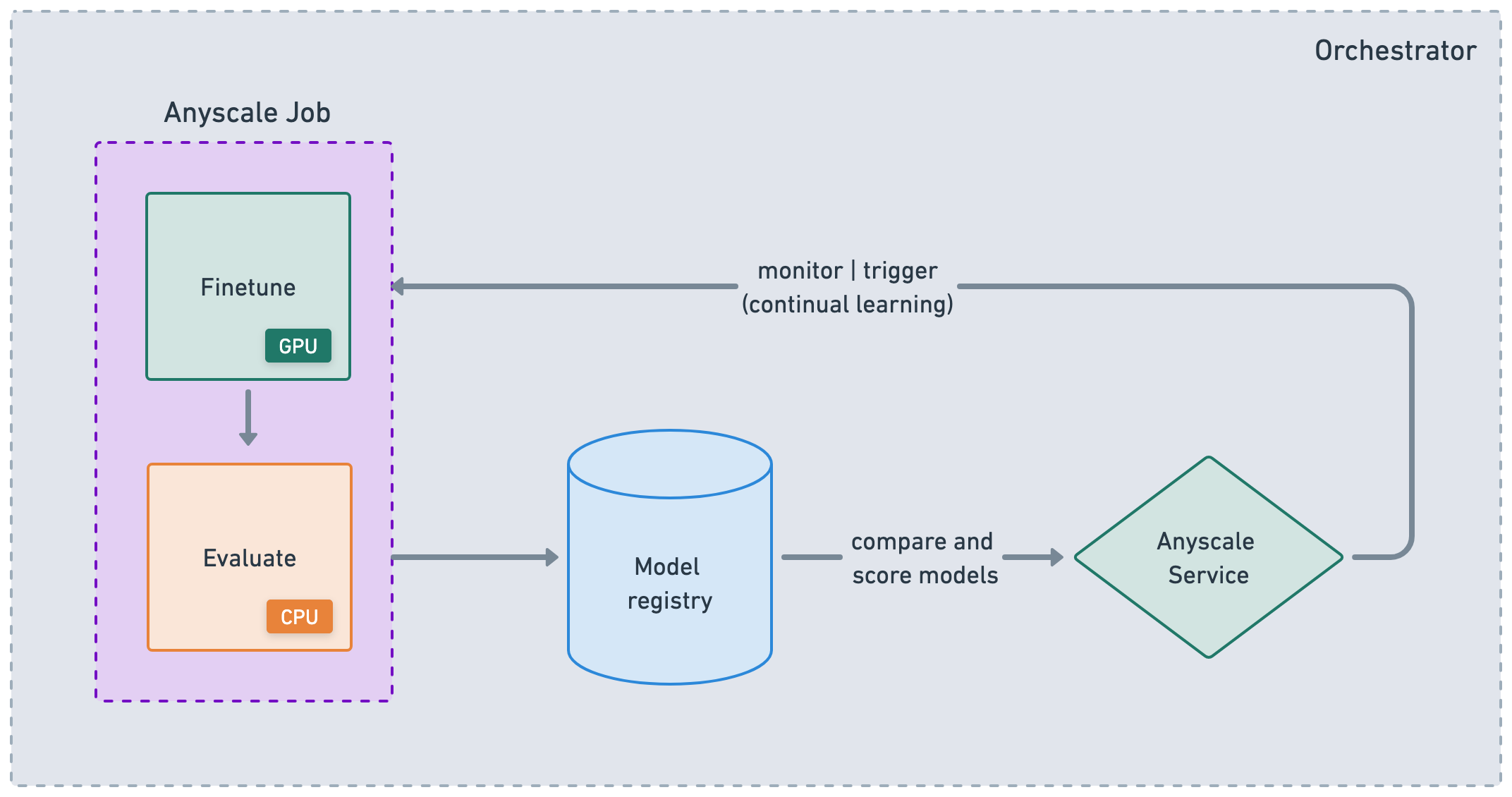
Anyscale Services offers a fault tolerant, scalable and optimized way to serve Ray Serve applications. See the API ref for more details. You can:
rollout and update services with canary deployment and zero-downtime upgrades.
monitor services through a dedicated service page, unified log viewer, tracing, set up alerts, etc.
scale a service with
num_replicas=autoand utilize replica compaction to consolidate nodes that are fractionally utilized.have head node fault tolerance. OSS Ray recovers from failed workers and replicas but not head node crashes.
serving multiple applications in a single Service
RayTurbo Serve on Anyscale has more capabilities on top of Ray Serve:
fast autoscaling and model loading to get services up and running even faster with 5x improvements even for LLMs
54% higher QPS and up-to 3x streaming tokens per second for high traffic serving use-cases with no proxy bottlenecks
replica compaction into fewer nodes where possible to reduce resource fragmentation and improve hardware utilization
zero-downtime incremental rollouts so the service is never interrupted
different environments for each service in a multi-serve application
multi availability-zone aware scheduling of Ray Serve replicas to provide higher redundancy to availability zone failures
Note:
This example uses a
containerfileto define dependencies, but you could easily use a pre-built image as well.You can specify the compute as a compute config or inline in a Service config file.
When you don’t specify compute and you launch from a workspace, the default is the compute configuration of the workspace.
# Production online service.
anyscale service deploy dist_xgboost.serve:xgboost_model --name=xgboost-breast_cancer_all_features \
--containerfile="${WORKING_DIR}/containerfile" \
--working-dir="${WORKING_DIR}" \
--exclude=""
Note that for this command to succeed, you need to configure MLflow to store the artifacts in storage that’s readable across clusters. Anyscale offers a variety of storage options that work out of the box, such as a default storage bucket, as well as automatically mounted network storage shared at the cluster, user, and cloud levels. You could also set up your own network mounts or storage buckets.
Running this command starts a service in production. In the process, Anyscale creates and saves a container image to enable fast starting this service in the future. The link to the endpoint and the bearer token appears in the logs. After the service is running remotely, you need to use the bearer token to query it. Here’s how you would modify the preceding requests code to use this token:
# Service specific config. Replace with your own values from the preceding logs.
base_url = "https://xgboost-breast-cancer-all-features-jgz99.cld-kvedzwag2qa8i5bj.s.anyscaleuserdata.com"
token = "tXhmYYY7qMbrb1ToO9_J3n5_kD7ym7Nirs8djtip7P0"
# Requests config.
path = "/"
full_url = f"{base_url}{path}"
headers = {"Authorization": f"Bearer {token}"}
prediction = requests.post(url, json=sample_input, headers=headers).json()
Don’t forget to stop the service once it’s no longer needed:
anyscale service terminate --name e2e-xgboost
While Anyscale Jobs and Services are useful atomic concepts that help you productionize workloads, they’re also convenient for nodes in a larger ML DAG or CI/CD workflow. You can chain Jobs together, store results, and then serve the application with those artifacts. From there, you can trigger updates to the service and retrigger the Jobs based on events, time, etc. While you can use the Anyscale CLI to integrate with any orchestration platform, Anyscale does support some purpose-built integrations like Airflow and Prefect.